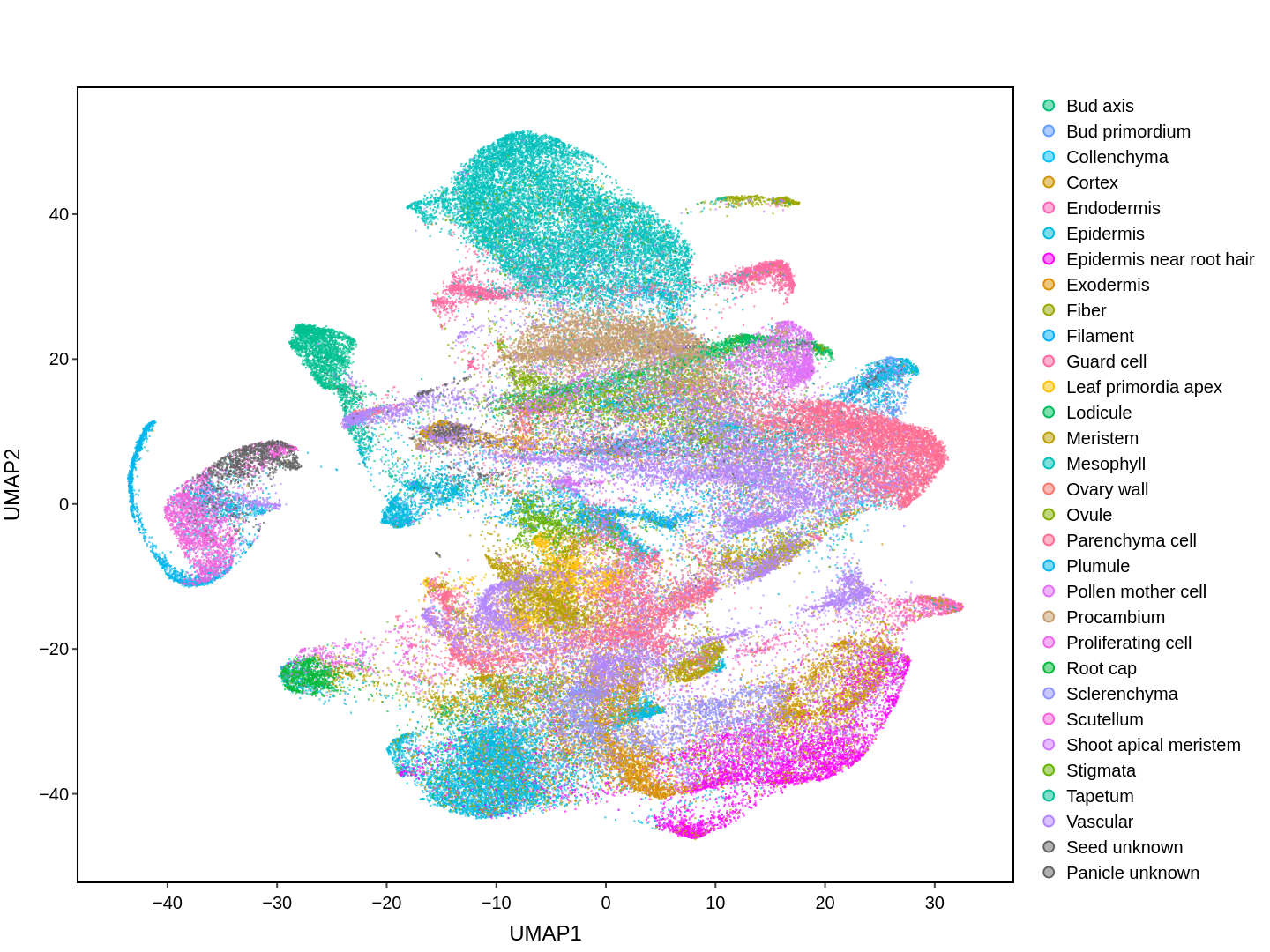
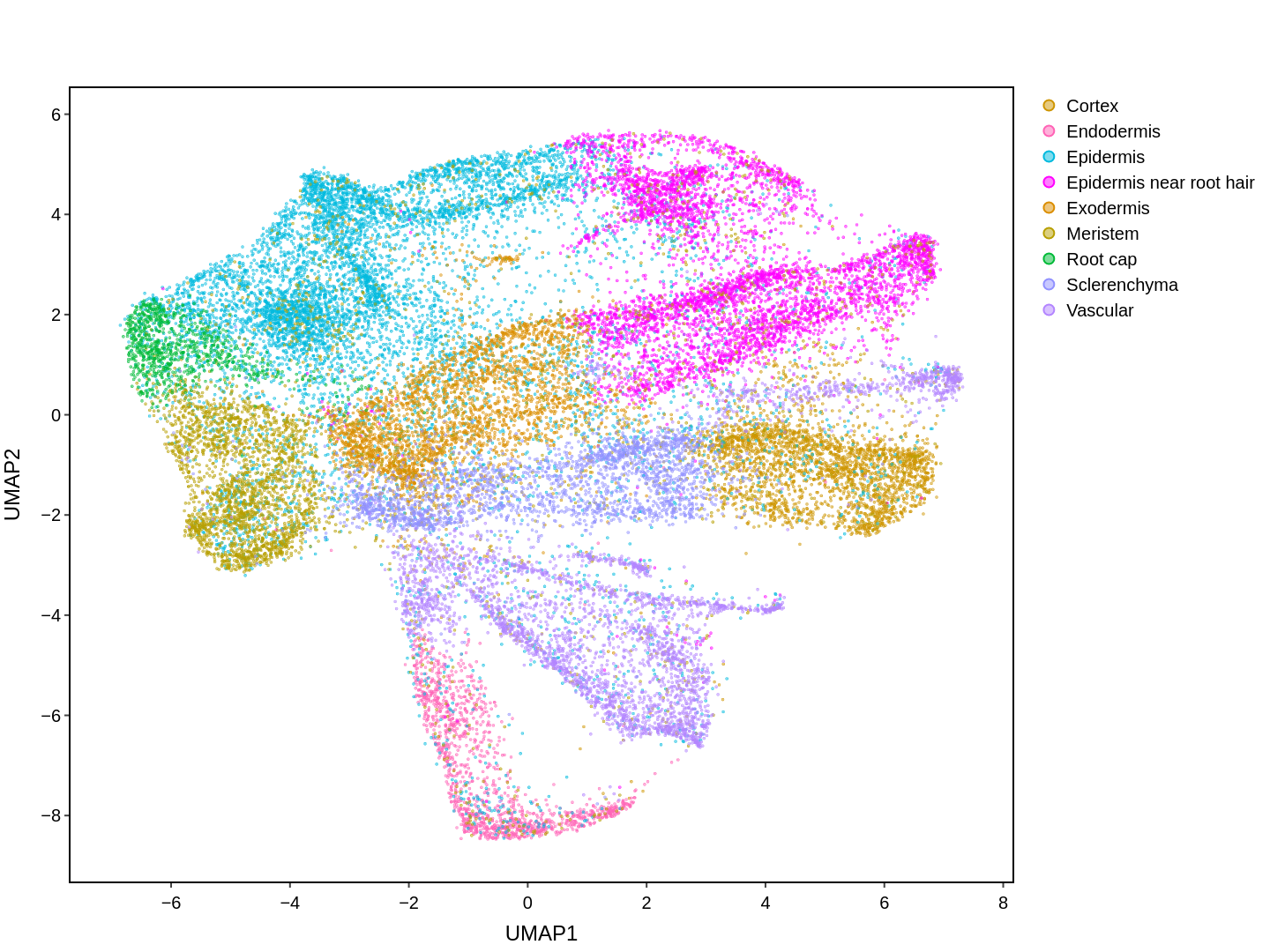
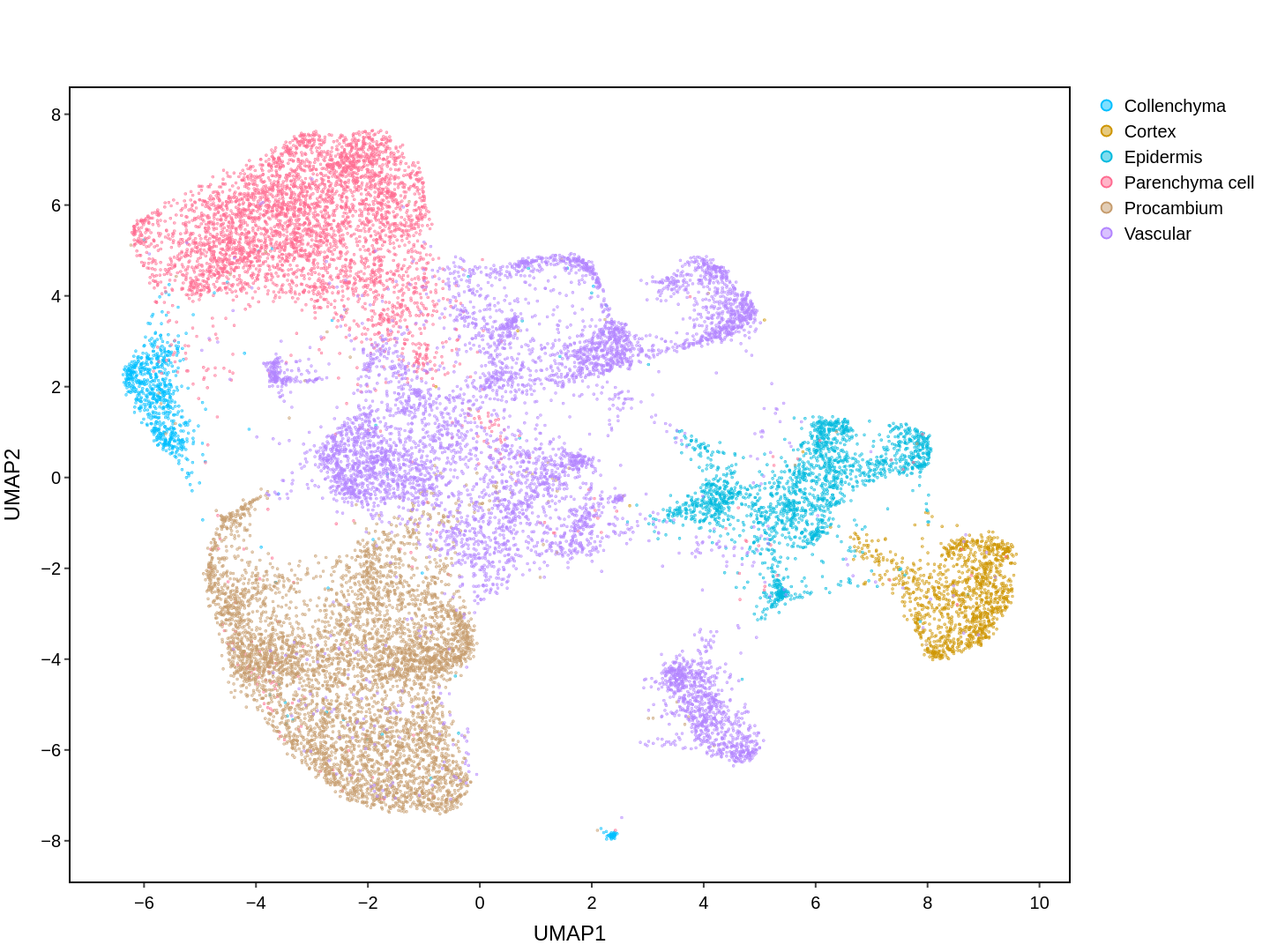
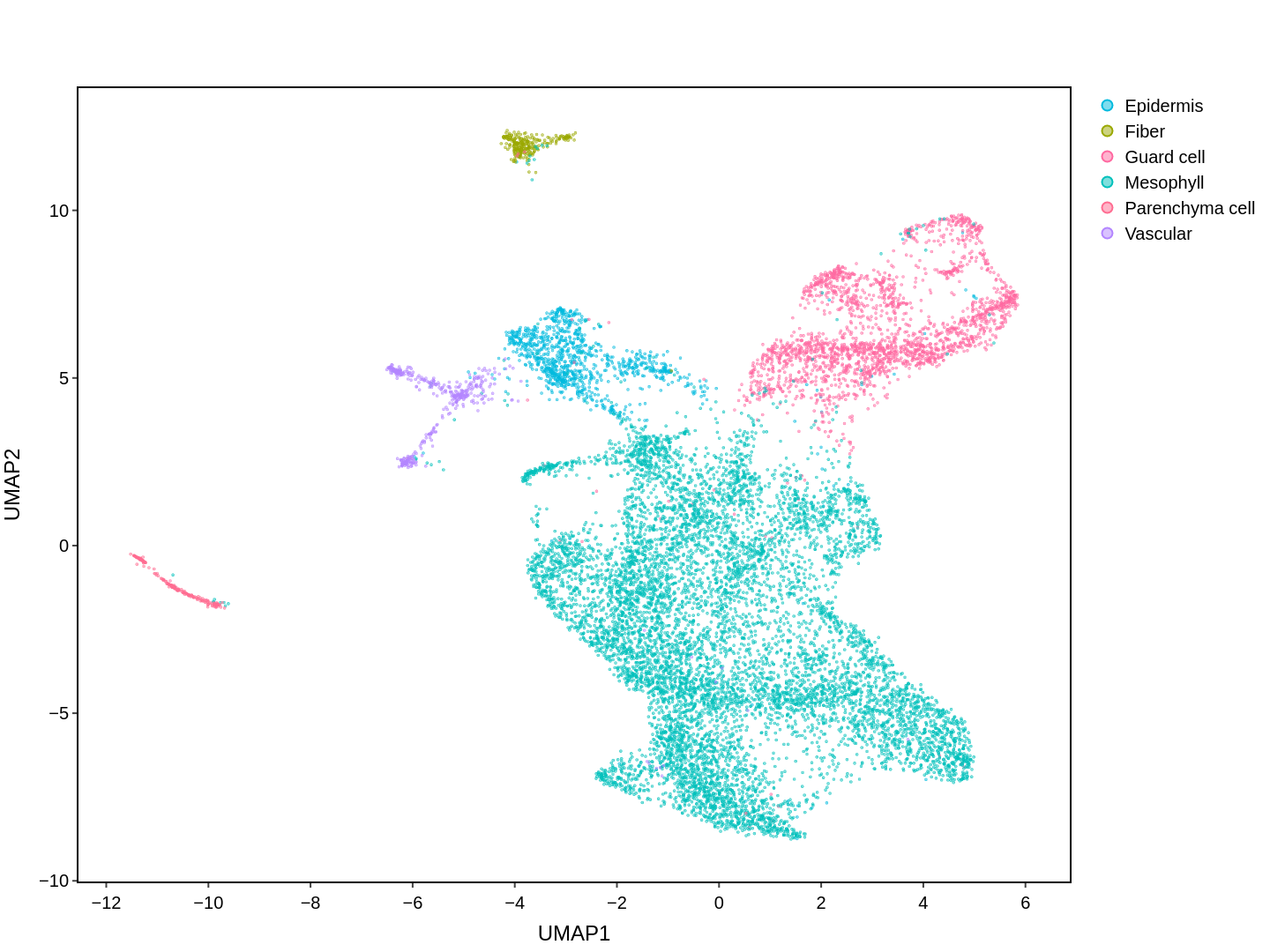
About Single Cell Multi-omics in Rice
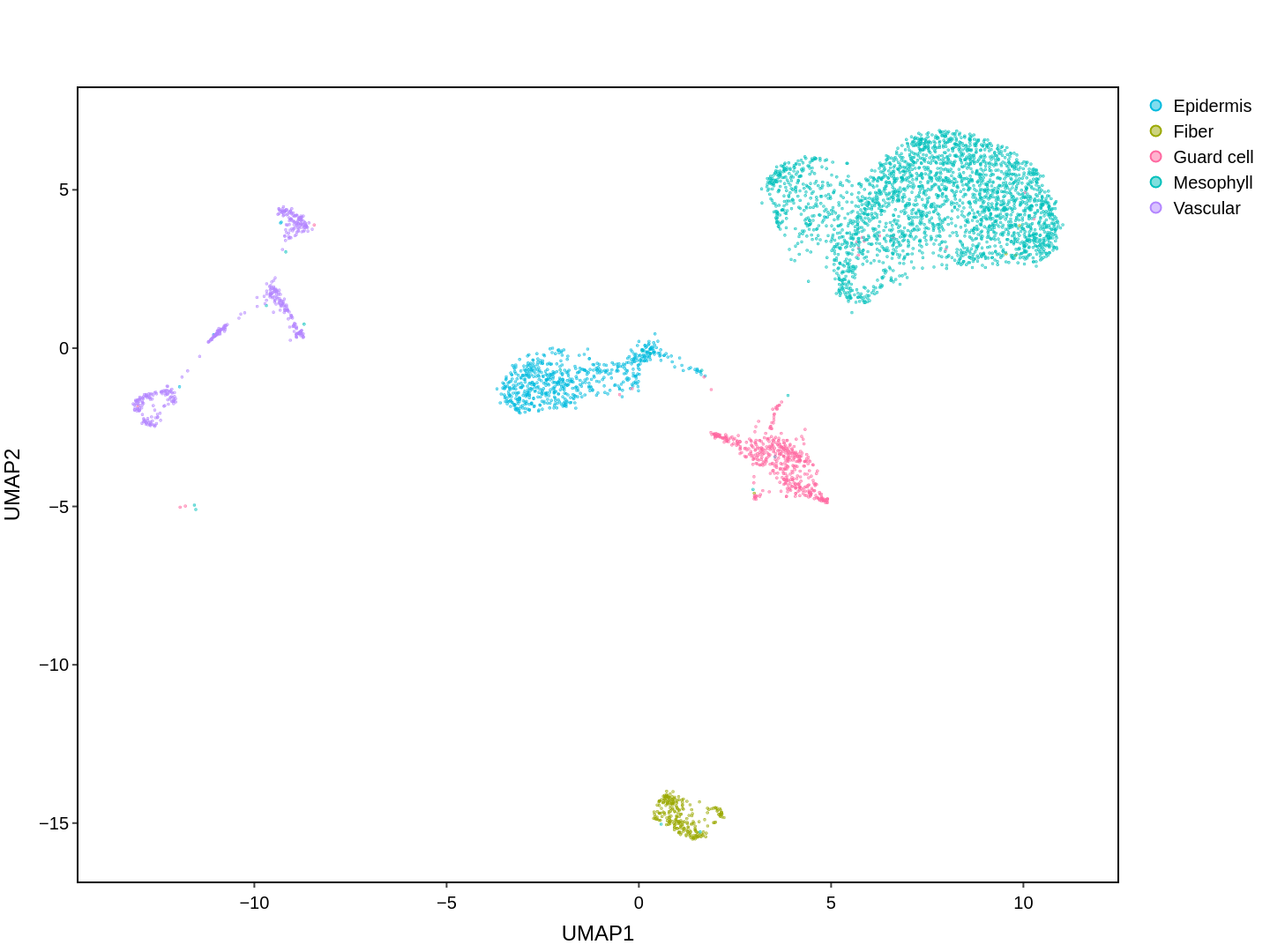
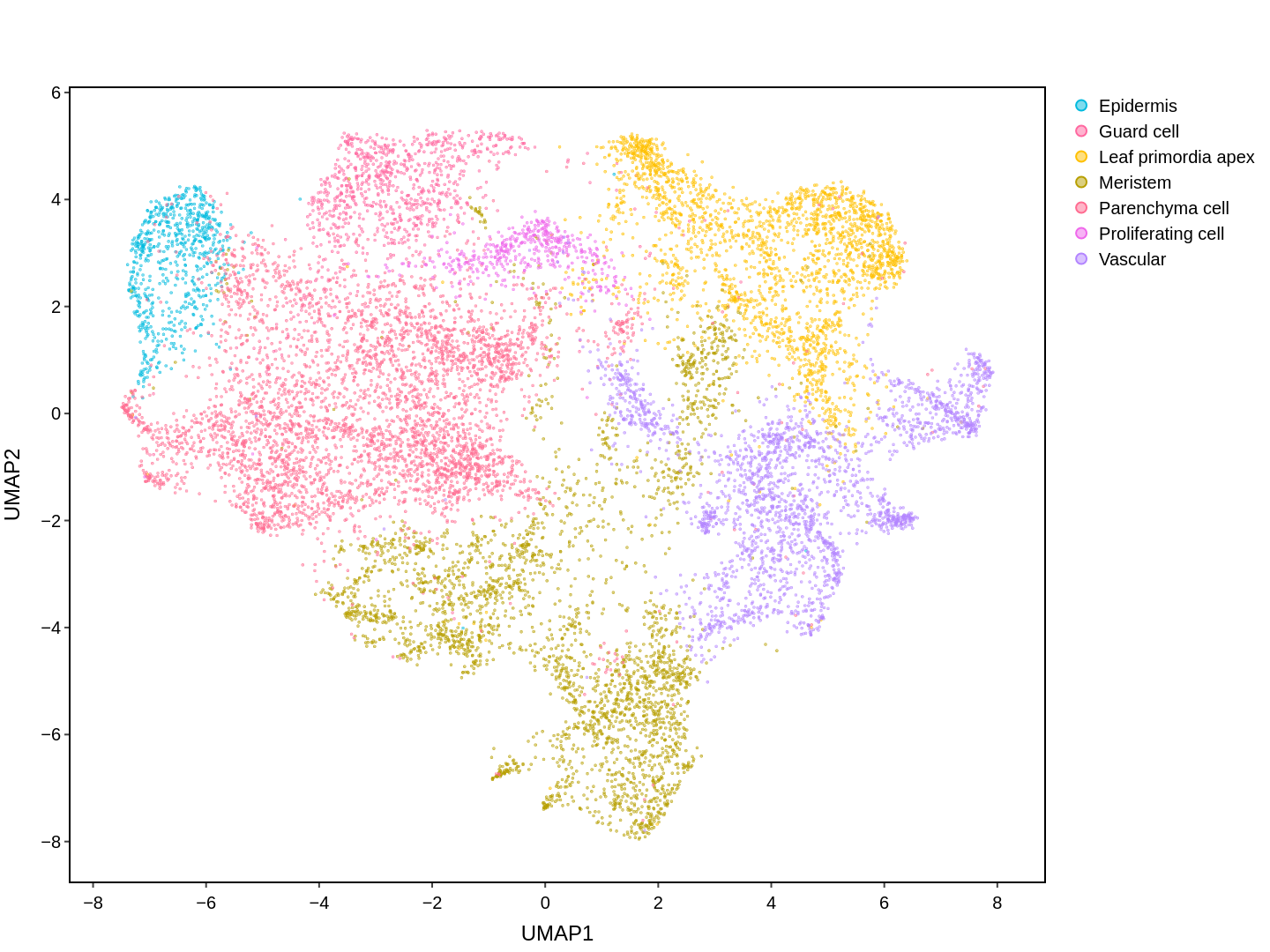
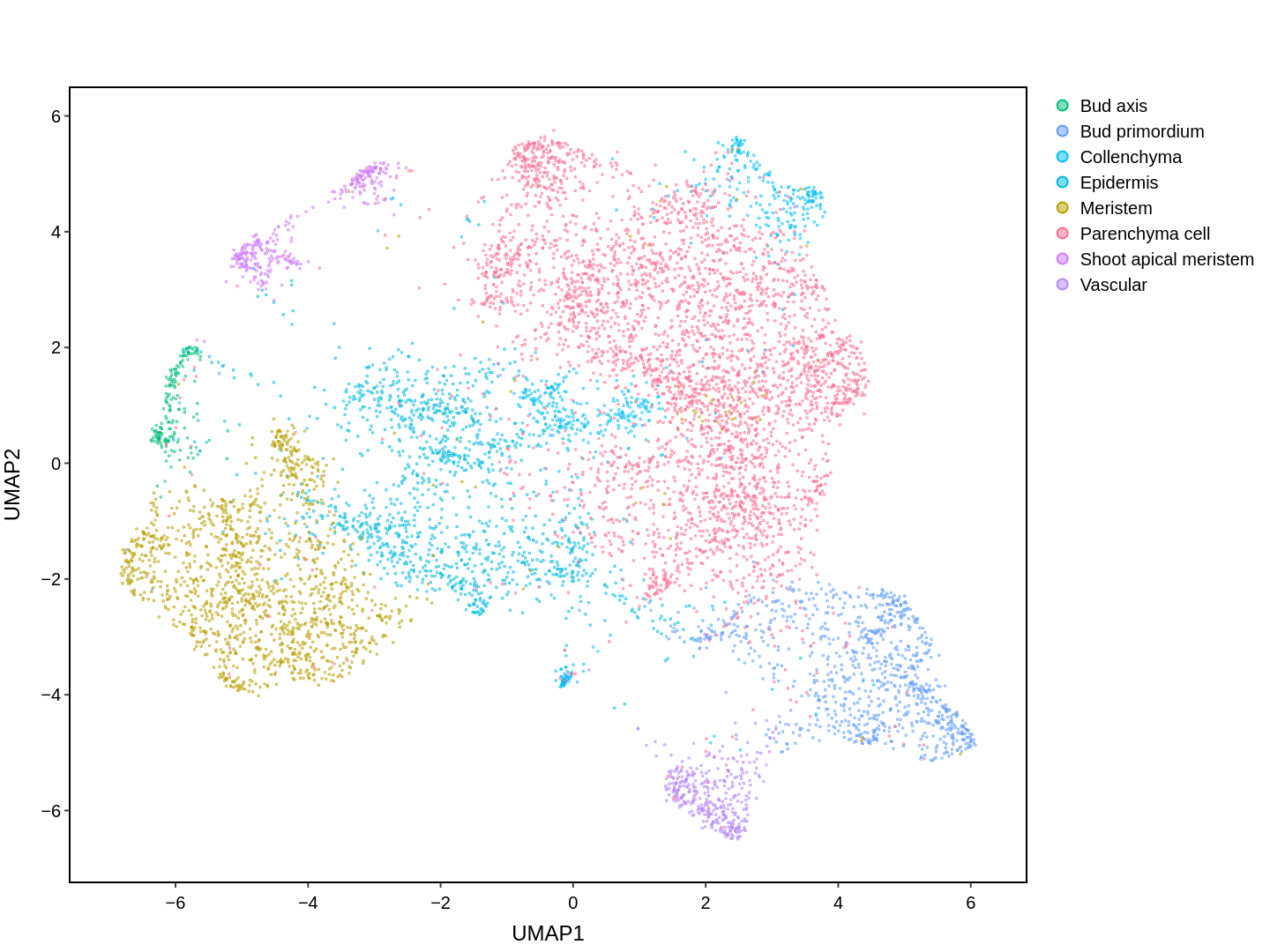
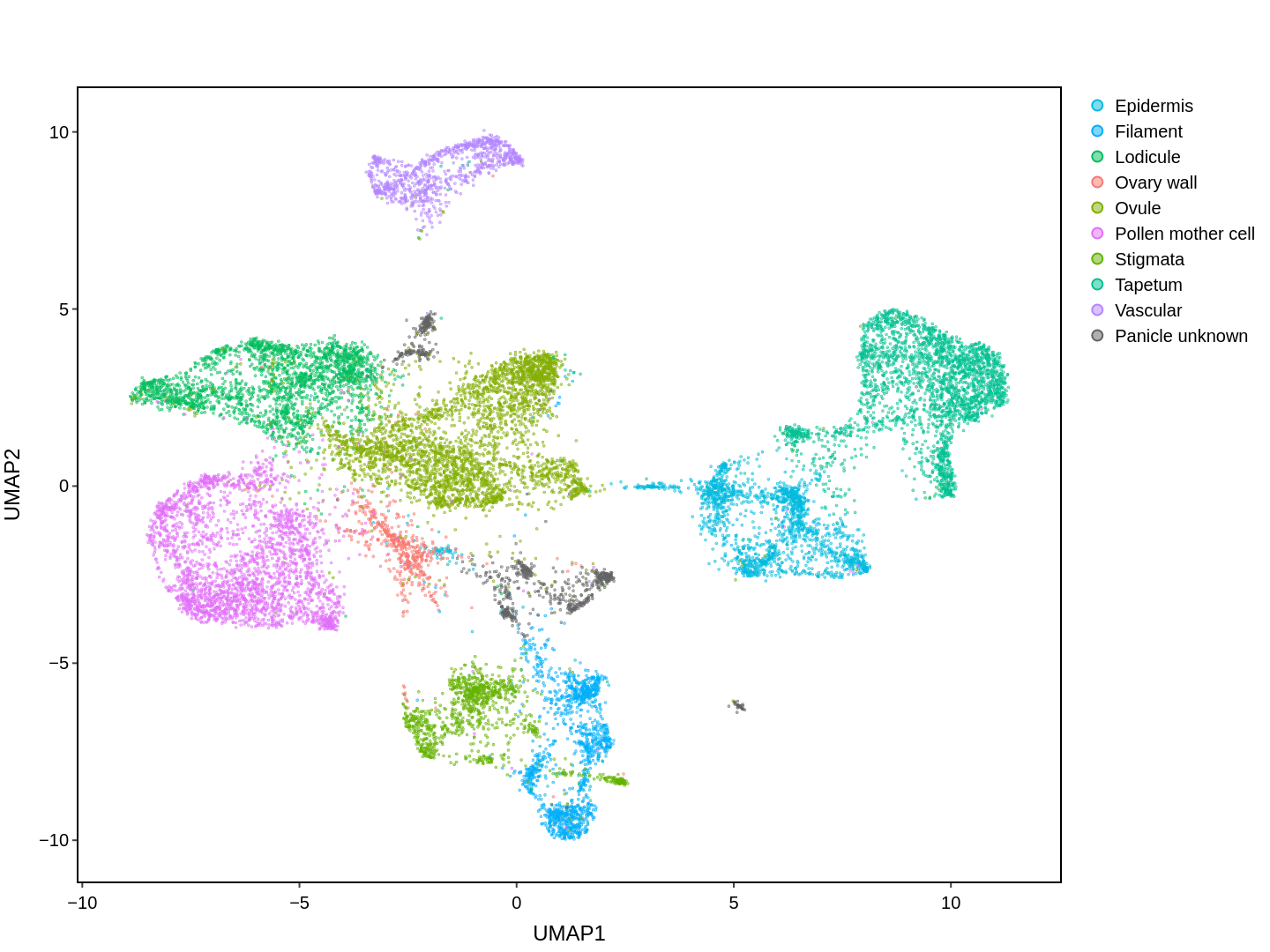
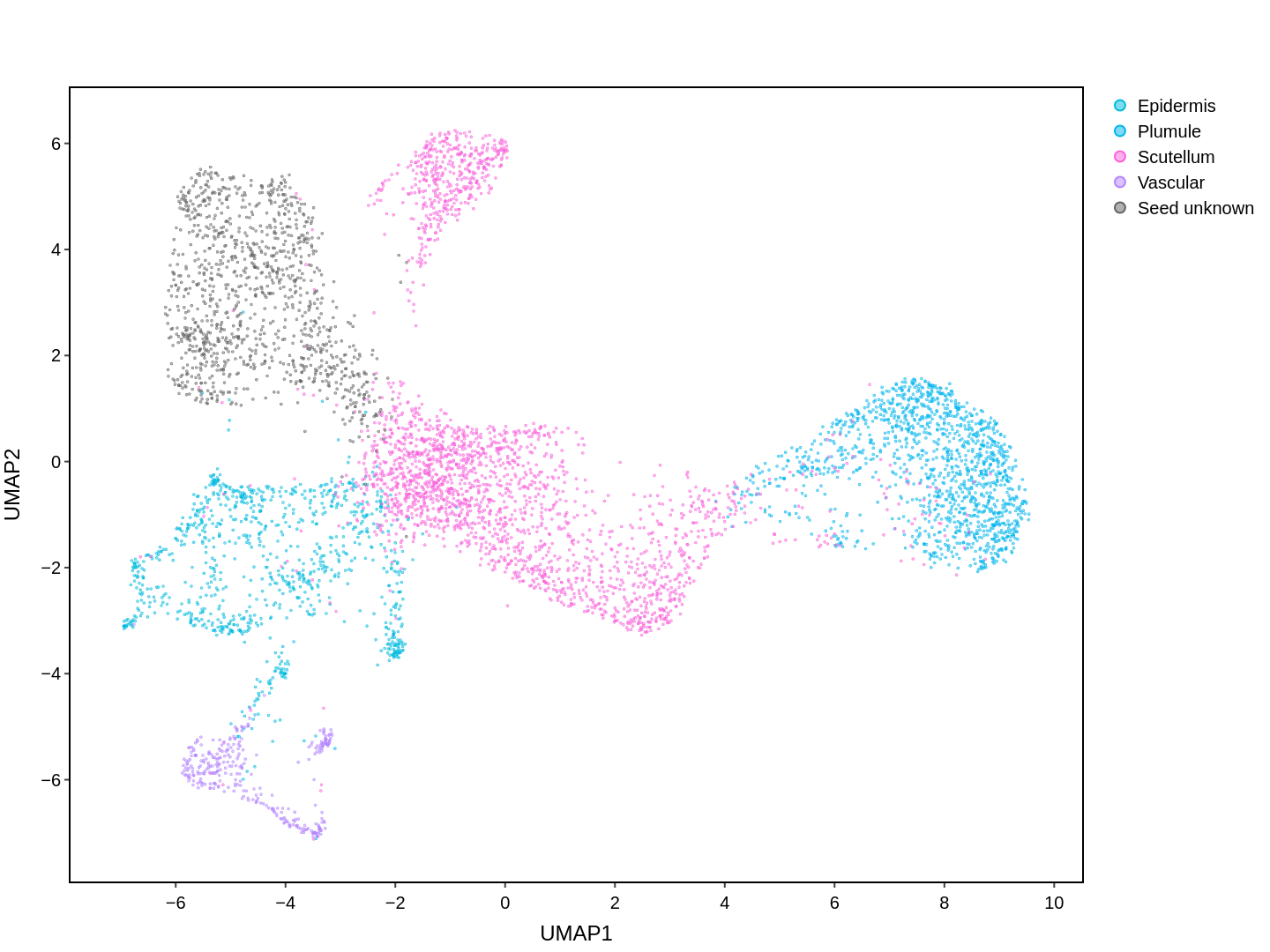
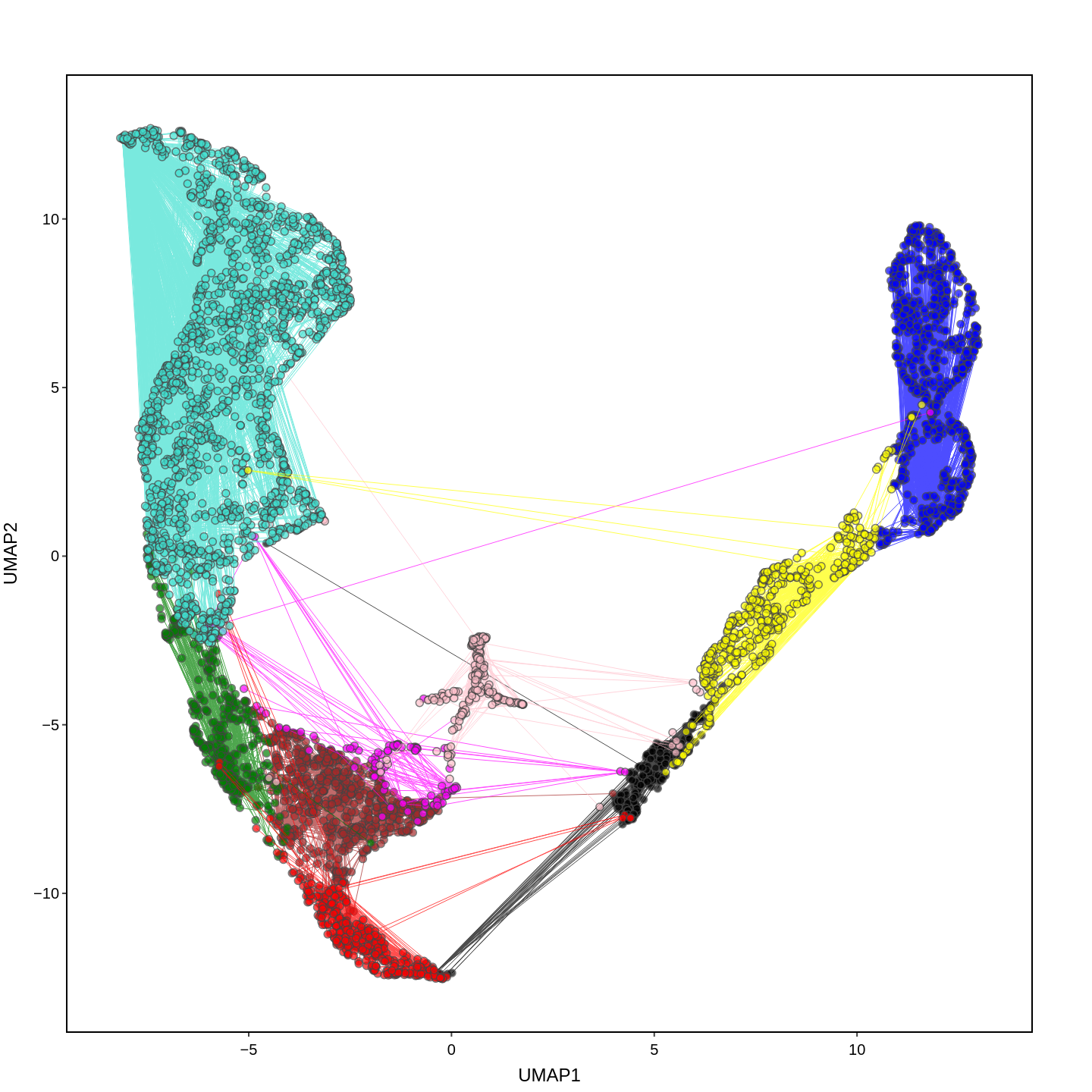
Cell functions across eucaryotes are driven by specific gene expression programs that are dependent on chromatin structure. Here, we report a single-cell multiomics atlas of rice, one of the major crop plants. We simultaneously profiled chromatin accessibility and RNA expression in 116,564 cells from eight organs. We annotated the majority of cell types and identified cell-type expression programs, and found a high correlation between RNA and chromatin accessibility across cells. We constructed single-cell gene regulatory networks and co-expression networks, which allowed us to identify the cell type-specific regulators RSR1, F3H and LTPL120 in rice development. Furthermore, our analysis revealed a correlation between cell type and agronomic traits of rice, and the conserved and divergent functions of cell type during evolution. In summary, this study not only offers a valuable single-cell multiomics resource, but also enriches our understanding of the intricate roles and molecular underpinnings of individual cell types in rice.
Tissues
Locus
You can search for the expression or chromatin accessibility status of each locus in each tissue or the whole plant.